Now Reading: Yerel Nükleozomlar Genom Organizasyonunun Temel İlkelerini Kodluyor
-
01
Yerel Nükleozomlar Genom Organizasyonunun Temel İlkelerini Kodluyor

Yerel Nükleozomlar Genom Organizasyonunun Temel İlkelerini Kodluyor
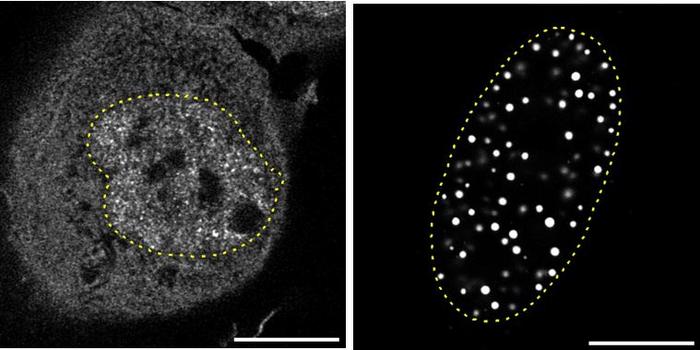
In a groundbreaking study published today in the prestigious journal Nature, researchers from the laboratory of Taekjip Ha at Boston Children’s Hospital have unveiled a novel and unexpected feature embedded within the fundamental units of our genome: nucleosomes. These tiny molecular structures, often likened to ‘tuna cans’ due to their compact, cylindrical form, have long been understood primarily as packaging elements that organize DNA into chromatin. However, the new findings reveal that nucleosomes carry an intrinsic physical code that actively guides the three-dimensional folding and spatial arrangement of the genome, tailoring chromatin architecture to meet the distinct functional demands of different tissues and organs.
This discovery represents a paradigm shift in molecular biology and genomics, providing compelling evidence that nucleosomes are not merely passive DNA holders but perform a highly nuanced and instructive role in genome organization. Through sophisticated experimental approaches and cutting-edge imaging techniques, the team demonstrated that the nucleosomes contain encoded mechanical and structural cues. These cues influence how chromatin fibers fold and interact within the cell nucleus, directing cells to develop specialized genome configurations essential for their specific physiological roles.
The ability of nucleosomes to dictate genome architecture on a nanoscale promises to illuminate longstanding questions about how cells maintain their identity and function amid the dynamic nuclear environment. Each tissue type requires unique gene expression patterns, and these patterns depend heavily on chromatin’s three-dimensional topology. The study suggests that nucleosomes act as architects of this spatial configuration, encoding information that predetermines how chromosomes loop, fold, and interact with regulatory elements such as enhancers and promoters.
Importantly, the insights gained from this research hold significant implications for understanding complex human diseases, particularly those marked by disruptions in genome architecture such as autoimmune disorders and cancer. Aberrant chromatin folding patterns have been implicated in gene misregulation underlying tumor progression and immune malfunction. By demonstrating that nucleosomes carry an inherent ‘blueprint’ for genome structure, the study opens up new avenues for therapeutic intervention that target the physical principles of chromatin organization.
The experimental findings stem from a multidisciplinary approach combining biophysical assays, single-molecule imaging, and advanced computational modeling. The researchers painstakingly isolated native nucleosomes from human cells and analyzed their folding behavior in vitro, revealing consistent, reproducible patterns that reflect intrinsic physical properties. These observations were confirmed by extensive simulations showing how nucleosome-encoded instructions translate into specific genome topologies observed in living cells.
Furthermore, the research sheds light on the longstanding debate about how epigenetic modifications influence genome architecture. While chemical modifications such as DNA methylation and histone tail alterations are known to modulate gene expression, this new evidence suggests physical structure encoded within nucleosomes adds an additional, heretofore underappreciated layer of regulatory complexity. This dual coding system — chemical and physical — provides a robust mechanism for fine-tuning genome function and responding adaptively to environmental cues.
The study’s revelations underscore the intricate relationship between genome structure and function, bridging gaps between genetics, molecular biology, and cellular physiology. The intrinsic nucleosome code helps explain how identical genomes across different cell types can produce vastly different gene expression profiles by reorganizing their three-dimensional frameworks. This finding challenges existing models that treated chromatin folding as solely dependent on external factors and epigenetic landscapes.
Moreover, this landmark discovery holds promise for future personalized medicine approaches. If nucleosomes possess physical codes that differ subtly between healthy and diseased states, then decoding these patterns could facilitate the development of diagnostic tools capable of detecting early structural abnormalities in chromatin. Such tools could revolutionize cancer prognosis and enable new strategies to restore normal genome architecture in disease conditions.
Taekjip Ha’s team also advocates that these findings could inform bioengineering tools designed to manipulate chromatin architecture deliberately. By harnessing the nucleosome’s inherent folding instructions, scientists might one day craft synthetic chromatin landscapes to control gene expression precisely, advancing gene therapy and regenerative medicine. This level of control would represent a monumental leap toward custom-designed cellular functions.
In addition to its transformative impact on human biology and medicine, the study profoundly affects fundamental science. It invites a reevaluation of the nucleosome’s role beyond traditional textbook definitions and inspires further research into how physical forces and structural codes interplay at the molecular level within the cell nucleus. Understanding this complexity is pivotal in decoding life’s blueprint in unprecedented detail.
The implications of this discovery extend even to evolutionary biology. If nucleosomes intrinsically encode genome organization, then changes in nucleosome properties over evolutionary timescales could have contributed to the diversification of genome architecture and, consequently, the emergence of complex multicellular life forms. This notion offers fertile ground for new hypotheses about the evolution of cellular complexity and specialization.
Ultimately, this pioneering research reveals that nucleosomes are more than passive DNA organizers; they are active participants encoding spatial and functional genome principles. By uncovering this hidden physical code, the study from Boston Children’s Hospital catalyzes a new era of inquiry into the molecular grammar of life itself. It sets the stage for revolutionary advancements in biology, medicine, and biotechnology, reshaping our understanding of how life’s instructions are encoded, interpreted, and executed within every human cell.
—
**Subject of Research**: Nucleosomes intrinsically encoding principles of genome organization and 3D chromatin architecture
**Article Title**: Native nucleosomes intrinsically encode genome organization principles
**News Publication Date**: 7-May-2025
**Web References**: Not provided
**Doi Referans**: 10.1038/s41586-025-08971-7
**Anahtar Kelimeler**: nucleosomes, genome architecture, chromatin folding, gene expression, molecular genetics, 3D genome organization, epigenetics, autoimmunity, cancer development, cellular function maintenance, physical code in nucleosomes, Taekjip Ha